Spatially resolved analysis in a cylindrical pore¶
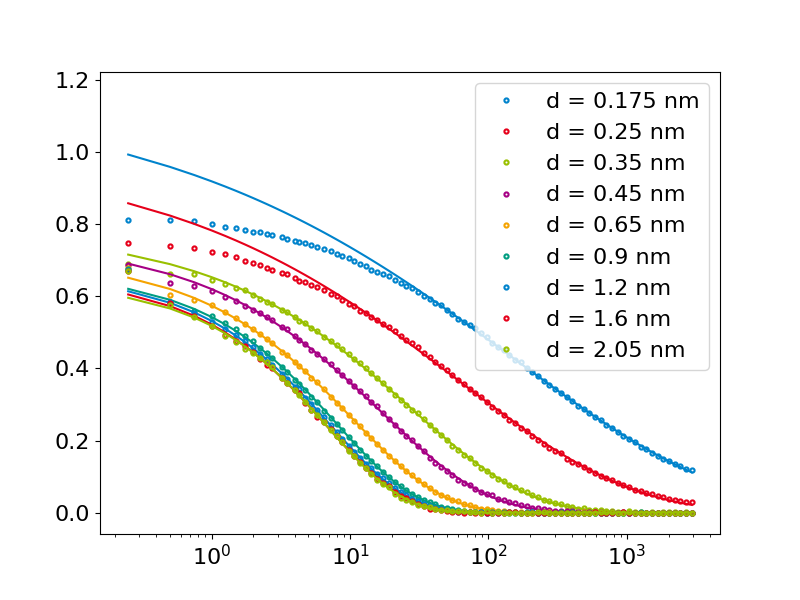
Calculate the spatially resolved ISF inside a cylindrical neutral water pore In this case the bins describe the shortest distance of an oxygen atom to any wall atom
import numpy as np
import matplotlib.pyplot as plt
import mdevaluate as md
import tudplot
from scipy import spatial
from scipy.optimize import curve_fit
#trajectory with index file
#TODO eine allgemeinere stelle?
traj = md.open('/data/robin/sim/nvt/12kwater/240_r25_0_NVT',
trajectory='nojump.xtc', index_file='indexSL.ndx',topology='*.gro')
#Liquid oxygens
LO = traj.subset(indices= traj.atoms.indices['LH2O'])
#Solid oxygens
SO = traj.subset(indices= traj.atoms.indices['SH2O'])
#Solid oxygens and bonded hydrogens
SW = traj.subset(residue_id = SO.atom_subset.residue_ids)
#TODO die folgenden beiden zusammen sind nochmal deutlich schneller als
#md.atom.distance_to_atoms, kannst du entweder in irgendeiner weise einbauen
#oder hier lassen, man muss aber auf thickness achten, dass das sinn macht
#adds periodic layers of the atoms
def pbc_points(points, box_vector, thickness=0, index=False, inclusive=True):
coordinates = np.copy(points)%box_vector
allcoordinates = np.copy(coordinates)
indices = np.tile(np.arange(len(points)),(27))
for x in range(-1, 2, 1):
for y in range(-1, 2, 1):
for z in range(-1, 2, 1):
vv = np.array([x, y, z], dtype=float)
if not (vv == 0).all() :
allcoordinates = np.concatenate((allcoordinates, coordinates + vv*box_vector), axis=0)
if thickness != 0:
mask = np.all(allcoordinates < box_vector+thickness, axis=1)
allcoordinates = allcoordinates[mask]
indices = indices[mask]
mask = np.all(allcoordinates > -thickness, axis=1)
allcoordinates = allcoordinates[mask]
indices = indices[mask]
if not inclusive:
allcoordinates = allcoordinates[len(points):]
indices = indices[len(points):]
if index:
return (allcoordinates, indices)
return allcoordinates
#fast calculation of shortest distance from one subset to another, uses pbc_points
def distance_to_atoms(ref, observed_atoms, box=None, thickness=0.5):
if box is not None:
start_coords = np.copy(observed_atoms)%box
all_frame_coords = pbc_points(ref, box, thickness = thickness)
else:
start_coords = np.copy(observed_atoms)
all_frame_coords = np.copy(ref)
tree = spatial.cKDTree(all_frame_coords)
first_neighbors = tree.query(start_coords)[0]
return first_neighbors
#this is used to reduce the number of wall atoms to those relevant, speeds up the rest
dist = distance_to_atoms(LO[0], SW[0], np.diag(LO[0].box))
wall_atoms = SW.atom_subset.indices[0]
wall_atoms = wall_atoms[dist < 0.35]
SW = traj.subset(indices = wall_atoms)
from functools import partial
func = partial(md.correlation.isf, q=22.7)
#selector function to choose liquid oxygens with a certain distance to wall atoms
def selector_func(coords, lindices, windices, dmin, dmax):
lcoords = coords[lindices]
wcoords = coords[windices]
dist = distance_to_atoms(wcoords, lcoords,box=np.diag(coords.box))
#radial distance to pore center to ignore molecules that entered the wall
rad = np.sum((lcoords[:,:2]-np.diag(coords.box)[:2]/2)**2,axis=1)**.5
return lindices[(dist >= dmin) & (dist < dmax) & (rad < 2.7)]
#calculate the shifted correlation for several bins
#bin positions are roughly the average of the limits
bins = np.array([0.15,0.2,0.3,0.4,0.5,0.8,1.0,1.4,1.8,2.3])
binpos = (bins[1:]+bins[:-1])/2
S = np.empty(len(bins)-1, dtype='object')
for i in range(len(bins)-1):
selector = partial(selector_func,lindices=LO.atom_subset.indices[0],
windices=SW.atom_subset.indices[0],dmin=bins[i],
dmax = bins[i+1])
t, S[i] = md.correlation.shifted_correlation(
func, traj,segments=50, skip=0.1,average=True,
correlation=md.correlation.subensemble_correlation(selector),
description=str(bins[i])+','+str(bins[i+1]))
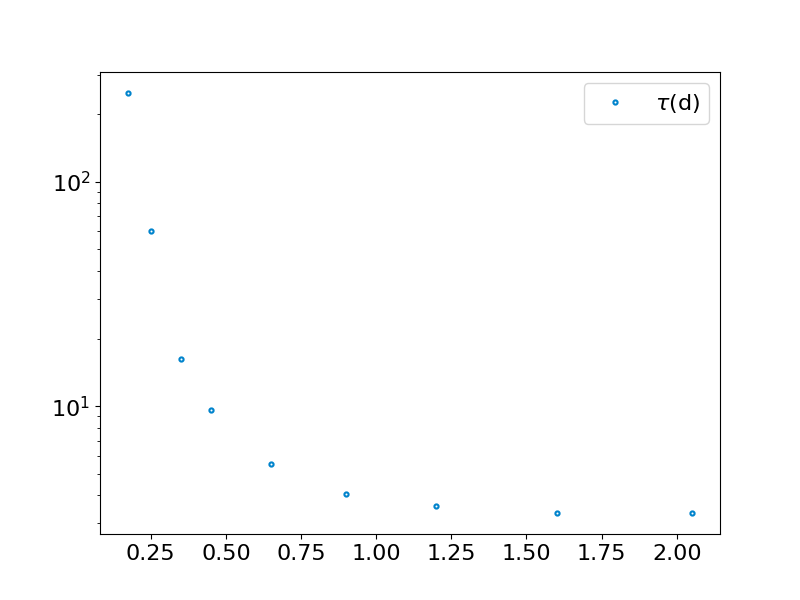
taus = np.zeros(len(S))
tudplot.activate()
plt.figure()
for i,s in enumerate(S):
pl = plt.plot(t, s, '.', label='d = ' + str(binpos[i]) + ' nm')
#only includes the relevant data for 1/e fitting
mask = s < 0.6
fit, cov = curve_fit(md.functions.kww, t[mask], s[mask],
p0=[1.0,t[t>1/np.e][-1],0.5])
taus[i] = md.functions.kww_1e(*fit)
plt.plot(t, md.functions.kww(t, *fit), c=pl[0].get_color())
plt.xscale('log')
plt.legend()
#plt.show()
tudplot.activate()
plt.figure()
plt.plot(binpos, taus,'.',label=r'$\tau$(d)')
plt.yscale('log')
plt.legend()
#plt.show()
Total running time of the script: ( 3 minutes 37.584 seconds)